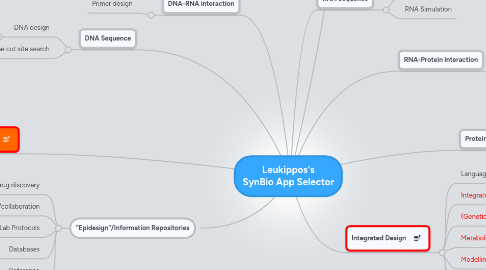
1. TO BE RECLASSIFIED:
1.1. Sequence analysis
1.2. Sequence alignment
1.3. Molecular structure (models)
2. DNA-RNA interaction
2.1. Primer design
3. DNA Sequence
3.1. DNA design
3.1.1. Parts design
3.1.2. Genome design
3.1.3. Sequence optimization
3.1.4. Sequence generation
3.2. Restriction enzyme cut site search
4. Consider excluding?
4.1. Education
4.1.1. Game
4.2. Hardware
4.2.1. Hardware drivers
4.2.2. Construction instructions
4.2.3. Laboratory automation
4.2.4. Robotics
4.2.5. Operation
4.3. Lab tools
4.3.1. E-lab notebooks
4.3.2. Image analysis
4.3.3. Calculator
4.4. Visualization
5. "Epidesign"/Information Repositories
5.1. Validating drug discovery
5.2. Data sharing/collaboration
5.2.1. Data sharing
5.2.2. Collaboration/information sharing
5.3. Lab Protocols
5.3.1. Protocol Generation
5.3.2. Physical assembly
5.3.3. Lab manual
5.3.4. Gibson assembly
5.4. Databases
5.4.1. BLAST search
5.4.2. Registry
5.4.3. Information resource
5.4.4. Repository
5.5. Reference
5.5.1. Part documentation
5.5.2. Literature
6. RNA Sequence
6.1. RNA design
6.2. RNA Simulation
7. RNA-Protein interaction
7.1. Codon optimization
7.2. Ribosome Binding Site
8. Protein analysis
8.1. Protein structure
8.2. Protein interaction prediction
9. Integrated Design
9.1. Language definition/Standards
9.2. Integrated workflow
9.2.1. Workflow management
9.2.2. Integrated design
9.2.3. Integrated framework
9.3. (Genetic) Circuit design
9.3.1. Circuit design and simulation
9.3.1.1. Biological network design
9.3.1.2. Network design
9.3.2. Circuit optimization
9.4. Metabolic engineering
9.4.1. Pathway generation
9.5. Modelling
9.5.1. Mathematical modelling
9.5.1.1. Matlab
9.6. Cell simulations
9.6.1. Bacteria Simulation
9.6.2. Simulations of Cell behavior
