1. Defintions
1.1. Antibiotic: antimicrobial substance made by a micro-organism
1.1.1. Fungi : Penicillium, Cephalosporium
1.1.2. Actinomycetes : Streptomyces, Micromonospora
1.1.3. (Soil bacteria) e.g streptomycin, gentamicin.
1.2. Antimicrobials : antibiotics AND chemically synthesised substances
1.2.1. e.g. sulphonamides, quinolones (ciprofloxacin etc.)
1.3. *Bactericidal = kill bacteria (some conditions)
1.4. Bacteriostatic = Stop growth/multiplication
1.5. *beta lactamase = enzyme which cleaves b lactam ring
1.5.1. (usually secreted in presence of antibiotics)
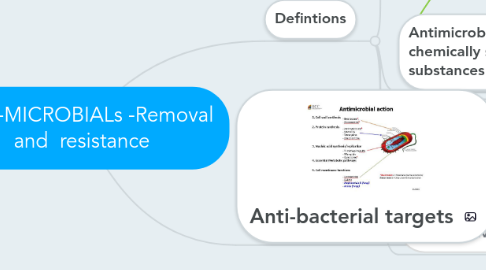
2. Anti-bacterial targets
2.1. 1. Cell wall synthesis
2.1.1. Beta lactams*,
2.1.1.1. Penicillin
2.1.1.1.1. Benzylpenicillin (penicillin G)
2.1.1.1.2. Phenoxymethylpenicillin (penicillin V)
2.1.1.1.3. Ampicillin/Amoxicillin
2.1.1.1.4. Flucloxacillin
2.1.1.1.5. Piperacillin
2.1.1.2. Cephalosporin
2.1.1.2.1. Info
2.1.1.2.2. LAME
2.1.1.2.3. Types
2.1.1.3. PBP
2.1.1.3.1. Catalysis of transpeptidation by PBP in cell wall
2.1.1.3.2. Transpeptidase crosslinks the peptidoglycan net in the cell wall of Gram-positive bacteria
2.1.1.3.3. B lactam MOA
2.1.1.4. Peptidoglycan (PGN)
2.1.1.5. Carbapenems
2.1.1.5.1. modified thienamycins from Streptomyces (no sulphur molecule)
2.1.1.5.2. for resistant Gram negative, Gram positive (not MRSA)
2.1.1.5.3. Injection only: meropenem, imipenem, ertapenem
2.1.1.6. Resistance
2.1.1.6.1. Removal of Drug:
2.1.1.6.2. Inactivation of Drug:
2.1.1.6.3. Alteration of Target:
2.1.1.6.4. Reduced intracellular penetration,
2.1.1.6.5. HOW?
2.1.2. Glycopeptide*
2.1.2.1. Vancomycin
2.1.2.2. Bind alanine subunits and prevent incorporation into PGN polymer
2.1.3. Bacitracin
2.1.3.1. interferes with PGN transfer to the cell wall by inhibiting dephosphorylation of lipid carrier
2.1.4. Cycloserine
2.1.4.1. structural analog of D-alanine and competes and inhibitd incorporation of D-alanine into peptidoglycan
2.2. 2. Protein synthesis
2.2.1. Acting on the bacterial ribosome
2.2.1.1. 50s
2.2.1.1.1. 23srRNA
2.2.1.1.2. 5srRNA
2.2.1.2. 30s
2.2.1.2.1. 16srRNA
2.2.2. Tetracyclines
2.2.2.1. Target: Gram positive and Gram negative
2.2.2.2. Side effects/toxicity
2.2.2.2.1. Deposited in growing teeth/ bone
2.2.2.2.2. Contraindicated in children and after 16th week of pregnancy
2.2.2.2.3. Photosensitisation
2.2.2.2.4. Hepatotoxicity
2.2.2.3. Resistance
2.2.2.3.1. Widespread resistance
2.2.2.3.2. mainly efflux and ribosomal protection
2.2.2.4. Tigecycline
2.2.2.4.1. Tetracycline derivative new broad spectrum agent
2.2.2.4.2. active against resistant Gram positive and negative bacteria
2.2.2.4.3. IV only
2.2.3. Macrolides
2.2.3.1. Source: Prototype erythomycin from bacterium Saccharopolyspora erythraea
2.2.3.2. Types
2.2.3.2.1. Erythromycin
2.2.3.2.2. Clarithromycin
2.2.3.2.3. Azithromycin
2.2.3.3. Target:
2.2.3.3.1. Chlamydia, Rickettsia, Legionella
2.2.3.3.2. Act on Gram positives, Mycoplasma
2.2.3.3.3. Azithromycin additional Gram negative activity
2.2.3.4. Administration: Oral and IV
2.2.3.5. Side effects:
2.2.3.5.1. Interact with statins, risk of myopathy
2.2.3.5.2. Immunomodulators
2.2.3.5.3. increase gastric motility
2.2.4. Aminoglycosides*
2.2.4.1. Natural products from soil bacteria
2.2.4.1.1. Actinomycetes : Streptomyces, Micromonospora
2.2.4.1.2. 1990s reduced use as fluoroquinolones
2.2.4.2. Types
2.2.4.2.1. Streptomycin,
2.2.4.2.2. Gentamicin
2.2.4.3. MOA:
2.2.4.3.1. bind 30s Ribosomal subunit
2.2.4.3.2. Misincorporation of Amino Acids into elongating peptides
2.2.4.3.3. Uptake requires Cytochrome mediated membrane potential
2.2.5. Oxazolidinone
2.2.5.1. Linezolid
2.2.5.1.1. TB
2.3. 3. Nucleic acid synthesis/replication
2.3.1. Synthetic antibacterials (types)
2.3.1.1. Rifampicin
2.3.1.2. -Trimethoprim/sulfa
2.3.1.3. Quinolones*
2.3.1.4. Oxazolidinones (Linezolid)
2.3.1.4.1. Source
2.3.1.4.2. MOA
2.3.1.4.3. Side Effects
2.3.1.4.4. Target
2.3.1.4.5. Admin
2.3.2. Inhibition of DNA replication
2.3.2.1. Quinolones (topoisomerase IV and DNA gyrase)
2.3.2.1.1. Source
2.3.2.1.2. MOA
2.3.2.1.3. Side Effects
2.3.2.1.4. Target
2.3.3. Inhibitors of RNA polymerase (mRNA synthesis)
2.3.3.1. Rifampicin
2.3.3.1.1. Source
2.3.3.1.2. MOA
2.3.3.1.3. Side Effects
2.3.3.1.4. Target
2.3.4. Antimetabolites inhibiting precursor synthesis
2.3.4.1. Antifolates (Sulphonamides, trimethoprim)
2.3.4.1.1. Bacteria make, but do not take up folate (B9)
2.3.4.2. Metronidazole (anaerobic infections only)
2.3.4.2.1. Source
2.3.4.2.2. MOA
2.3.4.2.3. Side Effects
2.3.4.2.4. Target
2.4. 4. Essential Metabolic pathways
2.5. 5. Cell membrane functions
2.5.1. Cell Membrane
2.5.1.1. Polymyxin
2.5.1.1.1. Gram Negative
2.5.1.1.2. Topical due to cell membrane issue
2.5.2. Lipopeptides
2.5.2.1. Daptomycin
2.5.2.1.1. Source
2.5.2.1.2. MOA
2.5.2.1.3. Target
2.5.2.1.4. Side Effects
2.5.2.1.5. IV only – reserve agent
2.5.3. Colistin
2.5.4. Azoles (Fungi)
2.5.5. Amphotericin B (Fungi)
2.6. Resistance
2.6.1. Removal of Drug:
2.6.1.1. Gram negatives, Outer membrane
2.6.2. Inactivation of Drug:
2.6.2.1. Gram neg and Gram pos
2.6.2.1.1. (bacterial beta lactamase enzyme hydrolyses the drug)
2.6.3. Alteration of Target:
2.6.3.1. Gram positives (MRSA, PBP2a)
2.6.4. Reduced intracellular penetration,
2.6.5. HOW?
2.6.5.1. Local selection of mutation
2.6.5.1.1. Chromosome
2.6.5.2. Horizontal transfer of resistance
2.6.5.2.1. Plasmid (conjugate,plasmid spread)
2.6.5.2.2. Transposon (gene spread)
2.6.5.2.3. Phage?
2.6.5.3. Resistance genes preceded therapeutic antibiotics
2.6.5.3.1. Present in environment e.g. soil.
2.6.5.3.2. Antibiotics seen as a subset of small inter-bacterial signalling molecules
2.6.5.4. therapeutic use
2.6.5.4.1. important selecting force
2.6.5.4.2. interpersonal spread of resistant bacteria is significant
2.6.5.4.3. The more an antimicrobial is used, the more selection pressure is exerted for the spread of resistance genes/resistant organisms
2.6.5.4.4. Indiscriminant use of antimicrobials risks rendering some common pathogens untreatable
